- +1 858 909 0079
- +1 858 909 0057
- [email protected]
- +1 858 909 0079
- [email protected]

Affinity chromatography, also known as bio-selective adsorption, uses specific binding affinity between two compounds. In this technology, a specific ligand is immobilized to an inert support matrix so that molecules with specific affinity to the ligand connect to it when a sample is passed through it. After washing away nonspecific molecules in the sample, the bound molecule(s) can be easily eluted from the ligand linked to the matrix. Affinity chromatography uses a protein’s biological structure to purify it, resulting in high capacity, selectivity, and resolution. As a result, affinity chromatography is frequently used for simple purification.
Affinity separation ligands include:
●
Small chemical molecules can attach to protein-binding sites
●
Inorganic metals that form coordination compounds with specific amino acids in proteins
●
Hydrophobic compounds that can bind nonpolar spaces in biomolecules
●
Proteins have particular binding sites that can interact with other proteins
●
Antibodies can be programmed to target any biomolecule via their antigen-binding sites
The use of immobilized affinity ligands to purify biomolecules has evolved beyond chromatographic applications with beaded agarose resins (still the most common). Magnetic beads have significant advantages over non-magnetic bead technologies. They thus see increasing use in various areas of life-sciences research and development, including drug discovery, biomedicine, bioassay development, diagnostics, genomics, and proteomics. This is due to the numerous benefits of magnetic beads, such as their ease of use, rapid experimental protocols, adaptability, and ease of use for high-throughput automated and miniaturized processing, such as high-throughput screening, as well as the potential for scaling due to the availability of massive homogenous biomagnetic separation equipment. Magnetic affinity and ion exchange separations have been used successfully in various domains, including molecular biology, biochemistry, immunochemistry, enzymology, analytical chemistry, and environmental chemistry.
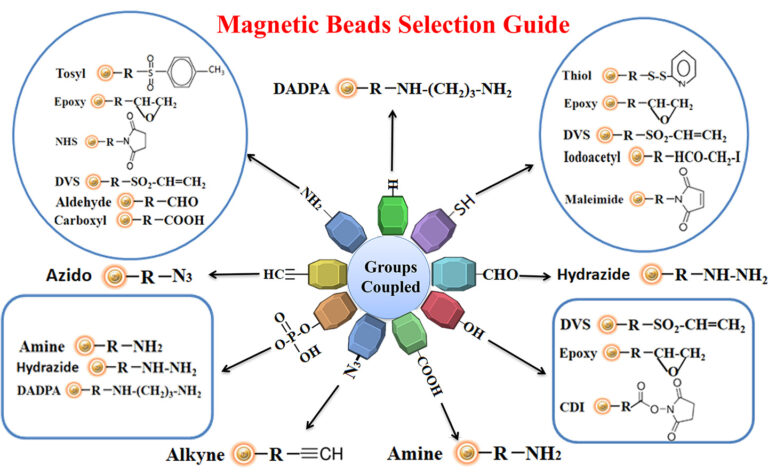
Making ligand-specific affinity magnetic beads requires methods to covalently conjugate ligands to appropriate magnetic beads. Immobilization and conjugation refer to the covalent attachment of ligands to magnetic beads through their common chemical groups. This method prepares the matrices by activating reactive chemical compounds toward one or more of these functional groups.
These chemical groups should be easily activated, such as primary amines, sulfhydryls, aldehydes, carboxylic acids, etc. The activated matrices then can conjugate antigens through a covalent linkage, resulting in ligand immobilization.
The type of linkage created between the matrix and the immobilized ligand influences the affinity support’s performance in a variety of ways. For example, if the linkage obstructs or negatively influences the structure of the immobilized ligand, its efficiency for affinity purification will be limited. A connection that allows the coupled ligand to leach from the matrix will contaminate the purified protein and reduce the affinity support’s useful life. Nonspecific binding can be caused by conjugation chemistry that puts a charged functional group into support by enhancing ion-exchange effects. A connection that changes the matrix’s structure can modify the flow and binding properties of the support.
Affinity ligands can be linked (immobilized) to the solid substance (stationary phase) in various ways.
The following general considerations apply to creating a successful magnetic affinity support matrix.
●
Linkers/spacers: Linkers are flexible molecules or stretches of molecules that link two molecules of interest together. The linkers’ property affects the affinity support’s performance in several ways.
●
Linker length. Suppose ligands are small peptide antigens or molecular. In that case, it is best to use a long arm linker (>10 atoms) attached to the support matrix since it can avoid steric hindrance (The ligand is inaccessible to the binding site) (Fig.). Linker length is less critical for large (e.g., protein) antigens since the antigen itself is an effective linker between the support matrix and the epitope.

●
Site-directed immobilization: Using a unique functional group for site-directed immobilization is beneficial for correct orientation, such as sulfhydryl on a single terminal cysteine in a peptide for a small antigen) and carbohydrate residues on the Fc side for antibodies.
●
Use a hydrophilic linker to immobilize antigen to the activated support to reduce background noise.
●
Use a cleavable linker to separate the target from the solid matrix easily.
Learn More
Immobilization chemistries
Affinity ligands can be linked (immobilized) to the solid substance (stationary phase) in various ways.
Reaction groups
The antigen is crosslinked with some carrier protein for the small antigen to make it immunogenic. For best results, we recommend that the antigen be immobilized to affinity purification using the same chemistry (e.g., reaction to primary amines, sulfhydryls, carboxylic acids, or aldehydes) for crosslinking the small antigen to the carrier protein. This way allows all epitopes to become available for antibody binding. Figure below summarizes the selection of chromatography resin selection for immobilizing antigens.
Pre-activated Magnetic Beads
Cleavable Magnetic Beads
Magnetic Beads Make Things Simple