- +1 858 909 0079
- +1 858 909 0057
- [email protected]
- +1 858 909 0079
- [email protected]

In the emerging field of proteomics, an essential step in understanding protein structure and function is to determine which proteins interact with each other, thereby identifying the relevant biological pathways. Pull-down assay is a type of affinity purification. It is similar to immunoprecipitation (IP) or co-immunoprecipitation (Co-IP), except that a bait protein is used to enrich prey proteins instead of an antibody. The Pull-down technique is widely used to detect or confirm interactions among multiple proteins predicted by other research techniques such as co-immunoprecipitation and analyze and quantify the specific isoforms of or active/inactive protein in the tested samples.
A pull-down assay utilizes a bait protein bound to beads to catch protein binding partners in a protein source that contains putative “prey” proteins, such as a cell lysate. The bait protein is usually fused with a tag, such as polyhistidine (His), glutathione-S-transferase (GS) biotin tag captured by Glutathione-, nickel chelate– or monomeric avidin magnetic beads, respectively. Pull-down assays involve isolating a protein complex by adsorbing the complex onto beads. Immobilized ligands on the beads bind specifically to a component of the complex via an affinity tag. In a typical pull-down assay, the process involves three steps: 1. The immobilized bait protein captures prey protein from a cell lysate during incubation. 2. Wash to eliminate the nonspecific binding. 3. Eluted the protein using competitive analytes or low pH or reducing buffers for SDS-PAGE or western blot analysis.
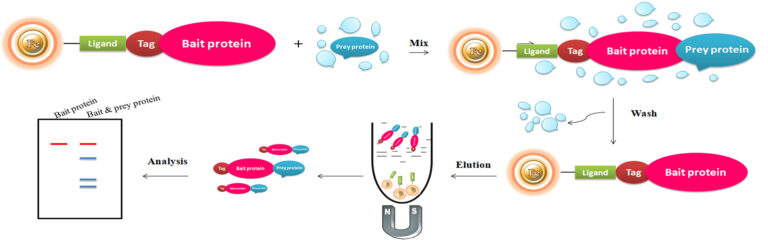
Bait proteins used for pull-down assays can be either ready-to-use purified proteins by linking an affinity tag or producing recombinant fusion-tagged proteins. In the representative example below, biotinylated protein is used to perform a pull-down assay.
Table 1 – Common Fusion Tags and Their Properties
Tag
Hexa-histidine
Size (kDa)
0.84
Ligand
Ni-IDA
Elution
Imidazole
Properties
Small size and easy to integrate into protein, simple purification process, mild elution condition
Tag
GST
Size (kDa)
26
Ligand
Glutathione
Elution
Glutathione
Properties
Simple purification process, mild elution condition
Tag
Biotin
Size (kDa)
0.244
Ligand
Monomeric avidin
Elution
Biotin
Properties
Small size, high specificity and affinity to avidin, simple purification process, mild elution condition
Size (kDa)
Ligand
Elution
Properties
Hexa-histidine
0.84
Ni-IDA
Imidazole
Small size and easy to integrate into protein, simple purification process, mild elution condition
GST
26
Glutathione
Glutathione
Simple purification process, mild elution condition
Biotin
0.244
Monomeric avidin
Biotin
Small size, high specificity and affinity to avidin, simple purification process, mild elution condition
A polyhistidine tag, also called 6xHis-tag, His-tagged, and His-tag is a versatile tool for purifying the highly purified recombinant protein from various expression systems, including bacterial, yeast, plant cell, and mammalian cells systems. The tag comprises six or more consecutive histidine amino acid residues positioned at either N or C terminus of a recombinant protein. Due to its small size, His-tag has several distinctive features, including less immunogenicity, hydrophilic nature, and versatility under native and denaturing conditions. Additionally, it is unnecessary to cleave the tag from the recombinant protein since it rarely perturbs the structure and function of its fusion protein. The purification principle of the His-tagged protein depends on immobilized Nickle magnetic beads and eluted the protein by imidazole.
Glutathione-S-transferase (GST) is a highly soluble and stable 26 kDa enzyme that catalyzes the protective mechanisms of Glutathione (GSH). Many eukaryotic proteins are produced as inclusion bodies (insoluble aggregated protein), a malfunctioning protein caused by misfolding, in prokaryotic expression systems such as E.coli. It is usually painful to refold the inclusion body into a functional protein. GST is widely used as a fusion partner that promotes greater expression and solubility of the desired protein by taking advantage of its high stability and solubility to overcome this problem. Moreover, GST has a high affinity toward reduced Glutathione, its natural substrate. Therefore, GST as an affinity tag becomes a versatile tool for single-step purification of active recombinant production in a prokaryotic expression system.
Glutathione is a short peptide (Glu-Cys-Gly) that displays a high affinity toward glutathione S-transferase (GST). When the Glutathione is immobilized in a chromatography matrix such as beaded agarose or magnetic beads, the matrix can precisely capture GST-tagged protein via the affinity interaction. The GST tag can be fused to either the C- or N-terminus of a protein by inserting DNA sequence coding for the protein of interest into commercial expression vectors. If desired, the protein of interest can be cleaved off the GST tag by site-specific protease. The protease site can be engineered between GST-tag and the protein of interest.
The interaction between avidin (or streptavidin) and biotin exhibits one of the highest known non-covalent interactions. Avidin, streptavidin, monomeric avidin, and their analogs have become powerful tools for probes and affinity ligands for various applications in biochemical assays, diagnosis, affinity purification, and drug delivery.
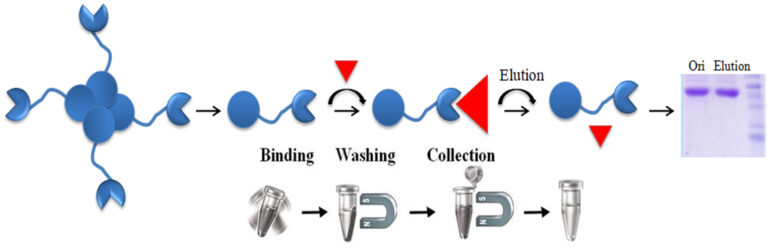
Materials Required
1.
Buffer
●
1x PBS Buffer (0.1 M sodium phosphate, 0.15 M sodium chloride; pH 7)
●
1x Regeneration Buffer (0.1 M Glycine/HCl, pH 2.8)
●
1x Blocking/Elution Buffer (2 mM D-biotin in PBS)
2.
Magnetic Rack (for manual operation)
Based on sample volume, the user can choose one of the following Magnetic Racks:
●
BcMag™ Magnetic Rack-2 for holding two individual 1.5 ml centrifuge tubes (Cat. No. MS-01);
●
BcMag™ Magnetic Rack-6 for holding six individual 1.5 ml centrifuge tubes (Cat. No. MS-02);
●
BcMag™ Magnetic Rack-24 for holding twenty-four individual 1.5-2.0 ml centrifuge tubes (Cat. No. MS-03);
●
BcMag™ Magnetic Rack-50 for holding one 50 ml centrifuge tube, one 15 ml centrifuge tube, and four individual 1.5 ml centrifuge tubes (Cat. No. MS-04);
●
BcMag™ Magnetic Rack-96 for holding a 96 ELISA plate or PCR plate (Cat. No. MS-05).
Protocol
The protocol can be adequately scaled up or down.
Note: Before purifying biotinylated proteins, peptides, and other molecules. The user should equilibrate all the reagents contained in the kit to room temperature and make 1x working solutions with double-distilled H2O.
1.
Gently shake the Magnetic Beads bottle until the magnetic beads are entirely suspended—transfer 50 µl beads to a fresh tube.
Note: Each user should empirically determine the optimal amount of beads to be used based on the amount of the biotinylated molecules in the crude sample. Too many magnetic beads will result in a higher background; too little will reduce the yield. We recommend 50 μl of the wholly suspended beads per 50 μg of biotinylated molecules.
2.
3.
Wash the beads with four bead volumes of 1x PBS buffer as described in step 2.
4.
Add three bead volumes of 1x Blocking / Elution Buffer, mix well by vortex, and incubate at room temperature for 5 minutes. Place the tube on the magnetic Rack for 1minute. Remove the supernatant while the tube remains on the Rack.
5.
Add six bead volumes of 1x Regeneration Buffer, mix well by vortex, and place the tube on the magnetic Rack for 1minute. Remove the supernatant while the tube remains on the Rack.
6.
Add four bead volumes of 1x PBS Buffer and wash the beads as described in step A.2. The beads are ready to use.
Note: Use the beads immediately, or binding capacity will dramatically reduce the binding capacity.
7.
Add biotinylated molecules-containing sample, mix well by pipetting and incubate at room temperature for 30-60 minutes with gentle rotation.
8.
9.
Add one bead volume of Blocking/Elution Buffer, mix well by pipetting several times, and incubate at room temperature for 5-10 minutes to elute the bound biotinylated molecule from the magnetic beads.
1.
Kim SY, Hakoshima T. GST Pull-Down Assay to Measure Complex Formations. Methods Mol Biol. 2019;1893:273-280. doi: 10.1007/978-1-4939-8910-2_20. PMID: 30565140.
2.
Einarson MB, Orlinick JR (2002) Identification of Protein-Protein Interactions with Glutathione S-Transferase Fusion Proteins. In: Protein-Protein Interactions: A Molecular Cloning Manual. Cold Spring Harbor (NY): Cold Spring Harbor Laboratory Press. pp 37–57.
3.
Einarson MB (2001) Detection of Protein-Protein Interactions Using the GST Fusion Protein Pulldown Technique. In: Molecular Cloning: A Laboratory Manual, 3rd Edition. Cold Spring Harbor (NY): Cold Spring Harbor Laboratory Press. pp 18.55–18.59.
4.
Vikis HG, Guan K-L (2004) Glutathione-S-Transferase-Fusion Based Assays for Studying Protein-Protein Interactions. In: Fu H (editor), Protein-Protein Interactions, Methods and Applications, Methods in Molecular Biology, 261. Totowa (NJ): Humana Press. pp 175–186.
5.
Louche A, Salcedo SP, Bigot S. Protein-Protein Interactions: Pull-Down Assays. Methods Mol Biol. 2017;1615:247-255. doi: 10.1007/978-1-4939-7033-9_20. PMID: 28667618.
Magnetic Beads Make Things Simple