- +1 858 909 0079
- +1 858 909 0057
- [email protected]
- +1 858 909 0079
- [email protected]

Immunoaffinity chromatography or immunoaffinity is a technique for separating an antigen or antibody from a biological sample based on the selective binding of the antibody to the antigen. Immunoaffinity chromatography utilizes an antibody or protein as a ligand immobilized onto a solid support matrix to capture the antigen or antibody. The target protein or antibody is later eluted and purified. It is a powerful technique for identifying, quantifying, or purifying antigens or antigens.
Antibodies (also known as immunoglobulin) are Y-shaped large glycoproteins produced by the immune system to identify and defend cells from foreign invading pathogenic bacteria or viruses. The antibody structure is shown in Figure below. The Y-shaped immunoglobulin consists of two identical heavy chains and light chains linked together by disulfide bonds. The two heavy chains are held together by disulfide bonds, and each heavy chain is connected to a light chain by a disulfide bond, forming a symmetrical “Y” molecule. The variable domains of heavy and light chains form the antigen-binding site.
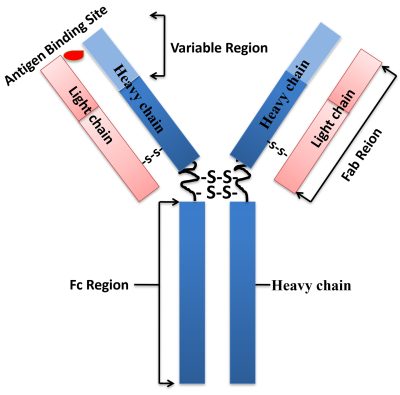
These antibodies are categorized into two main types: monoclonal (mAb) and polyclonal (pAbs). Polyclonal antibodies contain a heterologous mixture of IgGs produced by different B cells and recognize different epitopes (A small site on an antigen to which a complementary antibody may specifically bind precisely.) on the same antigen. Monoclonal antibodies are a homogenous antibody population produced from a single B-cell and recognize and bind to only one epitope on a single antigen. In mammals, antibodies are classified into five isotypes (IgG, IgM, IgA, IgD, and IgE) based on immunoglobulin structure.
Any molecule that induces the immune system to develop antibodies against it is referred to as an antigen. Antigens are typically proteins or polysaccharides with a large molecular weight. Antigens can also be polypeptides, lipids, nucleic acids, and various other substances. Immune responses can be elicited against smaller molecules known as haptens when they are chemically attached to a bigger carrier protein such as bovine serum albumin, keyhole limpet hemocyanin (KLH), or other synthetic matrices. Haptens can be compounds such as medicines, simple sugars, amino acids, short peptides, phospholipids, or triglycerides. As a result, given enough time, almost any foreign material will be recognized by the immune system and elicit particular antibody formation. However, this unique immune response is highly varied and is heavily influenced by antigen size, shape, and composition. Proteins or glycoproteins are thought to be the best antigens because of their potential to elicit a potent immune response; in other words, they are highly immunogenic.
Antigen-antibody interaction, also known as an antigen-antibody reaction, is a chemical interaction that occurs during an immune response between antibodies produced by B cells of white blood cells and antigens. Each antibody binds to a specific antigen, analogous to a lock and key contact. This lock and key affinity are helpful in biomedical engineering, biotechnology, and medicinal drug.
The antibody or antigen-specific affinity purification of antigen or antibodies involves two processes: Antibody or antigen immobilization to a solid support and antigen or antibody purification procedure from the biological samples. Making antibody- antigen-specific affinity supports requires methods to covalently conjugate antibodies or antigens to an appropriate insoluble support (matrix). Immobilization and conjugation refer to the covalent attachment of an antibody or antigen to a solid support through their common chemical groups. This method prepares the matrices by first activating with reactive chemical compounds toward one or more of these functional groups. These chemical groups should be easily activated, such as primary amines, sulfhydryls, aldehydes, carboxylic acids, etc. The activated matrices then can conjugate antibodies or antigens through a covalent linkage, resulting in ligand immobilization.
1.
Choice of solid supports
Although antibody or antigen-specific affinity purification of antibodies is efficient, most support matrices consist of the agarose resin or its derived column. The problems with their procedures in sample preparation are tedious, time-consuming, unable to handle multiple samples in a short time, and challenging to adapt to the automation system. New affinity resins with higher efficiency, homogeneity, and high throughput are required. Affinity ligands are now coupled to magnetic particles that facilitate more efficient capture of specific biomolecules.
Magnetic beads (particles) are an entirely different type of solid support matrices from beaded agarose or other porous resins. They are much smaller (typically 1-5 µm diameter), thus providing larger surface areas for a high density of ligand immobilization. The beads are manufactured using nanometer-scale superparamagnetic iron oxide as core and entirely encapsulated by a high purity silica shell, ensuring no leaching problems with the iron oxide. The excellent hydrophilic properties of the beads make them less nonspecific binding. Bioclone offers many activated functional groups and variable-length hydrophilic linkers for conjugating almost all antibodies or antigens. Additionally, the beads combine all the advantages of antibody or antigen-specific purification (low costs, simplicity, high specificity, and capacity) and magnetic properties to perform efficient manual or automatic quick high-throughput purification.
2.
Immobilization chemistry
The successful antibody or antigen-specific affinity purification of antigen or antibodies depends on the availability and effectiveness of the relevant epitope on the antigen to the binding site of the antibody and immobilization efficiency of the antigen to the affinity support matrix. The following general considerations apply when antigens are immobilized to solid affinity supports.
●
Linkers/spacers: Linkers are flexible molecules or stretch of molecules that link two molecules of interest together. The linkers’ property affects the affinity support’s performance in several ways.
●
Linker length. Suppose ligands are small peptide antigens or molecular. In that case, it is best to use a long arm linker (>10 atoms) attached to the support matrix since it can avoid the steric hindrance (The ligand is inaccessible to the binding site) (Fig.). Linker length is less critical for large (e.g., protein) antigens since the antigen itself is an effective linker between the support matrix and the epitope.

●
Site-directed immobilization: Using a unique functional group for site-directed immobilization is beneficial for correct orientation such as sulfhydryl on a single terminal cysteine in a peptide for a small antigen) and carbohydrate residues on the Fc side for antibody.
●
Use hydrophilic linker to immobilize antigen to activated support to reduce background noise.
●
Use a cleavable linker for easy separation of the target from the solid matrix.
Reaction groups
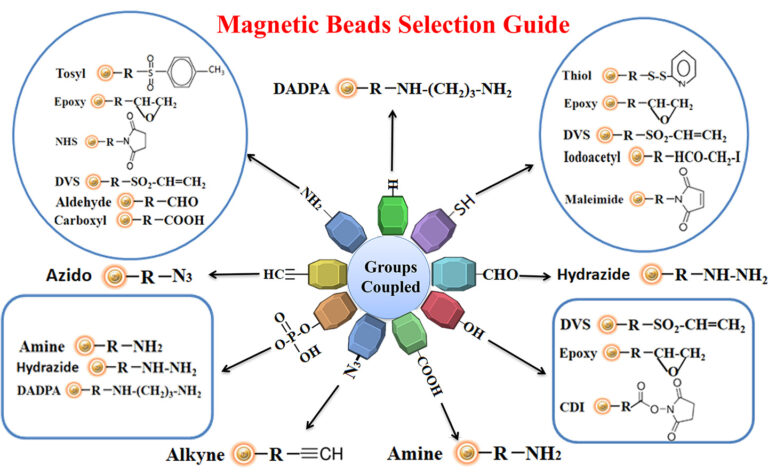
The antigen is crosslinked with some carrier protein for the small antigen to make it immunogenic. For best results, we recommend that the antigen be immobilized to affinity purification using the same chemistry (e.g., reaction to primary amines, sulfhydryls, carboxylic acids, or aldehydes) for crosslinking the small antigen to the carrier protein. This way allows all epitopes to become available for antibody binding. Figure below summarizes the selection of chromatography resin selection for immobilizing antigens.
Learn More
The interaction between avidin (or streptavidin) and biotin exhibits one of the highest known non-covalent interactions. Avidin, streptavidin, monomeric avidin, and their analogs have become powerful tools for probes and affinity ligands for various biochemical assays, diagnosis, affinity purification, and drug delivery applications.
Advantages and benefits of using biotin-binding protein systems
●
High affinity
●
High stability
The biotin-binding complex is exceptional stability and can resist Extreme pH, temperature (2 °C and 40 °C), harsh organic solvents, denaturing agents such as guanidinium chloride, detergents such as SDS and proteolytic enzymes.
●
Outstanding selectivity and specificity
The biotin and streptavidin or avidin interaction are highly specific, ensuring low nonspecific binding.
●
High sensitivity
High stability and specificity ensure high detection sensitivity.
●
Very flexibility
The small size and remarkable stability of biotin are exceptionally suitable for relatively easy incorporation into various molecules or specific locations in molecules with minimal perturbation to the structure and function of the conjugated biomolecules.
Applications
Compared to other non-covalent interactions, the biotin-binding system provides tremendous benefits. It has become a versatile platform for amplifying weak signals, flow cytometry, Western blotting, ELISA, FLISA, IHC, immunofluorescence microscopy, etc.
Learn More
Explore Products
Protein A, G, A/G, and L are native and recombinant proteins whose antibody-binding properties have been well characterized. The native proteins are replaced by the recombinant proteins produced in E.coli since the recombinant proteins have higher capacity, are highly robust, and have maximum specific antibody binding. The characteristics of these recombinant proteins are summarized in Table 1.
Table 1. Characteristics of Protein A, Protein G, Protein A/G, Protein L
Protein A
Protein G
Protein A/G
Protein L
Original Source
Staphylococcus aureus
Staphyl-ococcus aureus
Streptococcus spp.
(Group C and G)
Strepto-coccus spp.
(Group C and G)
Staphylococcus aureus & Streptococcus spp.
(Group C and G)
Staphyl-ococcus aureus & Strepto-coccus spp.
(Group C and G)
Peptostreptococcus magnus
Peptostr-eptoco-ccus magnus
Recombinant Size (E.coli)
Recombi-nant Size (E.coli)
33 kDa
22 kDa
50 kDa
38 kDa
Number of Ig Binding Domains
5
2
6
5
Ig-binding site
Protein A, Protein G, or Protein A/G binds with high affinity to the Fc portion of various classes and subclasses of immunoglobulins from various species by independent and separate binding sites. Protein L binds the different antibody isotypes (IgG, IgM, IgA, IgE, and IgD) through the interaction with the variable domain of the Ig kappa light chain with no interference with an antibody’s antigen-binding site. These proteins vary in their ability to bind to different subtypes and species antibodies. Therefore, choosing the antibody-binding proteins to match the corresponding antibody is essential. Refer to table 2 to select the antibody-binding protein that is best for your application.
Due to remarkable antibody binding characteristics, the proteins are widely used in antibody purification, immunoprecipitation (IP), chromatin immunoprecipitation (ChIP), immobilization or detection of immunoglobulins.
Learn More
Explore Products
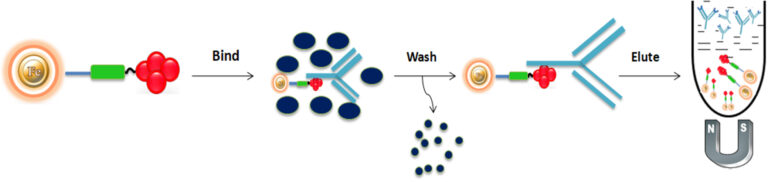
Generally, there are three steps (Bind-Wash-Elute) involving immunoaffinity purification.
●
Bind
Add the antigen or antibody-bound magnetic beads to the cell lysate. Mix and incubate at room temperature for 1-2 hours with continuous rotation to allow the antigen to bind to the immobilized ligand.
●
Wash
Wash the beads with phosphate-buffered saline (PBS) to remove non-bound serum components. Use a volume of wash buffer equivalent to 5 to 10 times the bead-bed volume.
●
Elute
Elute the antigen-antibody complex from the beads with acidic elution buffer (e.g., 0.1 M glycine-HCl, pH 2.8) and collect the supernatant.
●
Neutralize or exchange Buffer
Immediately neutralize the eluted antigen by adding 1/10th volume of 1 M Tris-HCl (pH 8.5) and mix well. Several analysis techniques can further characterize the antigen.
1.
Phillips, T. M. (1989). High-performance immunoaffinity chromatography. Adv. Chromatogr.29, 133–173.
2.
Weliky, N., Weetall, H. H., Gilden, R. V., and Campbell, D. H. (1964) Synthesis and use of some insoluble immunologically specific adsorbents. Immunochemistry1, 219–229.
3.
Kristiansen, T. (1976 ). Matrix-bound antigens and antibodies. Scand. J. Immunol.(Suppl.) 3, 19–27
4.
Santucci, A., Soldani, D., Lozzi, L., et al. (1988) High performance liquid chromatography immunoaffinity purification of antibodies and antibody fragments. J. Immunol. Methods114, 181–185.
5.
Muronetz, V. I., Sholukh, M., and Korpela, T. (2001) Use of protein-protein interactions in affinity chromatography. J. Biochem. Biophys. Methods49, 29–47.
6.
Hage, D. S. (1998) Survey of recent advances in analytical applications of immunoaffinity chromatography. J. Chromatogr.715, 3–28
7.
Hill, C. R., Kenney, A. C., and Goulding, L. (1987) The design, development and use of immunopurification reagents. Biotech. Forum4, 168–171.
8.
Dunbar, B. S. and Schwoebel, E. D. (1990) Preparation of polyclonal antibodies, in Methods in Enzymology (Deutscher, M. P., ed.), Academic, London, vol. 182, pp. 663–670.
Magnetic Beads Make Things Simple