- +1 858 909 0079
- +1 858 909 0057
- [email protected]
- +1 858 909 0079
- [email protected]

The dynamical interaction between RNAs and RNA-binding proteins (RBPs) plays essential roles in controlling gene expression programs, and several methods have been recently developed to map them. One of the powerful techniques to investigate the interaction is RNA Immunoprecipitation (RIP) assay. RNA Immunoprecipitation (RIP) is an antibody-based immunoprecipitation used to purify the mRNA and noncoding RNA associated with specific nuclear or cytoplasmic binding proteins from a cell lysate. RIP assay allows the rapid identification of endogenous RNAs bound to an RBP and monitors their dynamical changes in response to different metabolic and stress conditions. The purified RNA-protein complex can be characterized and analyzed by techniques including reverse transcription-polymerase chain reaction (RT-PCR), microarray analysis (RIP-chip), and next-generation sequence (RIP-Seq).
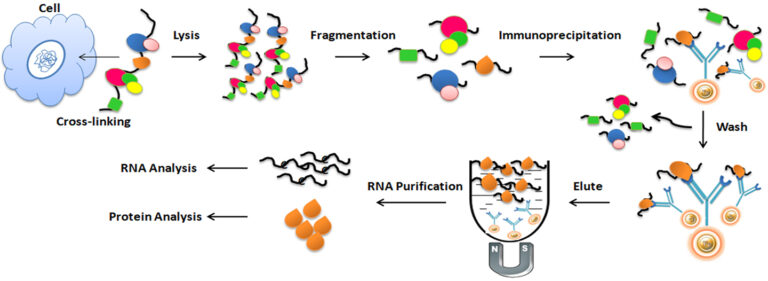
RNA Immunoprecipitation (RIP) assay involves five steps: crosslinking, fragmentation, immunoprecipitation, RNA Purification, and analysis of the enriched RNA.
1.
Crosslinking
Formaldehyde is usually used to crosslink protein and RNA at a particular site. Optimizing the crosslinking reaction time is necessary since excessive crosslinking may reduce antigen accessibility and sonication efficiency.
2.
Fragmentation
For efficient immunoprecipitation, sonicating chromatin into small fragments is necessary. Again, optimizing the sonication condition is essential for individual applications.
3.
Immunoprecipitation
Selecting an appropriate antibody for RIP assays is critical for its success. The best choice of antibody used for RIP assays should be a RIP- or RIP-seq-validated antibody. The antibody should have high specificity and a strong affinity for the protein of interest. In addition, it is also vital to carefully choose an appropriate antibody binding protein, such as protein A/G or protein L, which matches the selected antibody.
Bead Choice
●
Magnetic vs. Agarose Beads
Magnetic beads (particles) are an entirely different type of solid support matrices from beaded agarose or other porous resins. They are much smaller (typically 1-5 µm diameter), thus providing larger surface areas for a high density of ligand immobilization. They are easy separation and visibility in the tube, leading to less loss of material and highly reproducible results. In addition, the beads are compatible with the exited automation system. Agarose beads have a relatively higher capacity for binding due to their porous nature. However, they have higher nonspecific binding.
●
Protein A vs. Protein G vs. Protein A/G vs. Protein L
Protein A/G has a broader binding range than Protein A or Protein G individually. While Protein L binds a broader range of antibody classes than Protein A or G since the heavy chain is not involved in the binding interaction. Protein L binds to most Ig classes, single-chain antibody fragments (scFv), and Fab fragments.
The antibody to the protein of interest or the antibody immobilized to the protein A/G magnetic beads is then mixed and incubated with the cell lysate mixture at 4oC to form bead/antibody/protein/RNA complexes. The beads are then extensively washed to remove the nonspecifically binding using a magnet rack. The antibody/protein/RNA complex is finally eluted from the beads and collected for further RNA purification.
Protocols for Immunoprecipitation
4.
RNA Purification
Before RNA purification, it is necessary to use proteinase K and DNase to reverse the crosslinks and digest all the associated proteins, antibodies, and DNA. A commercial RNA purification kit can purify the RNA.
5.
Analysis of the enriched RNA
The purified RNA is then analyzed by reverse transcription-polymerase chain reaction (RT-PCR), microarray analysis (RIP-chip), and next-generation sequence (RIP-Seq) to identify and quantify the sequences.
One-Step vs. Multistep PCR Purification
RNA Immunoprecipitation followed by sequencing analysis (RIP-Seq) is a high-throughput method to investigate genome-wide RNA transcripts interacting with a protein or protein complex. Library construction for next-generation sequence (NGS) typically involves a multistep process including end-repair, adapter-ligation, size selection, gel purification, and PCR using primers specific to the sequencing platform. PCR purification is time-consuming in constructing a library, especially for handling multiple samples.
Unlike the traditional “Bind-Wash-Elute” multistep PCR purification method, One-Step PCR Cleanup Kit uses only one step, one minute, and one tube to finish the purification. The beads enable 96 samples to be processed simultaneously in less than 10 minutes. The beads can remove the impurities (e.g., excess primer, dimer, adapter, salt, detergent, dNTPs, and enzyme) and remove the different size RNA fragments by adjusting processing time, buffer pH, and detergent concentration.
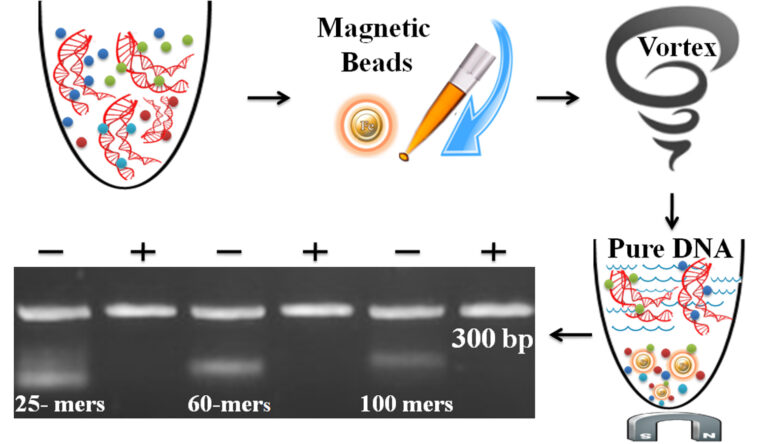
Magnetic Beads Make Things Simple