- +1 858 909 0079
- +1 858 909 0057
- [email protected]
- +1 858 909 0079
- [email protected]

Products
Components
Shipping conditions: At ambient temperature
Components
BcMag™ Oligo-dT Magnetic Beads
1x Binding / Lysis Buffer
10x Washing Buffer I
10x Washing Buffer II
Elution Buffer
Storage
4°C
4°C
4°C
4°C
4°C
Cat. No. MMS101 (50 Preps)
2 ml
10 ml
5 ml
5 ml
0.5 ml
Cat. No. MMS102 (100 Preps)
4.0 ml
Shipping conditions: At ambient temperature
Handling and Storage: Store the kit components according to the table Above on arrival.
Product Specificities
Composition
Magnetic beads linked with oligo-dT (25)
Bead Size
~1μm diameter
Number of Beads
~1.7 x 108 beads (1μm beads) /mg
Stability
pH -6-10
Magnetization
~40-45 EMU/g
Type of Magnetization
Superparamagnetic
Effective Density
2.5 g/ml
Concentration
10 mg/ml in TE Buffer
Binding Capacity
10 µg mRNA / ml of beads
Storage
Store Magnetic beads at 4°C, Buffers at room temperature upon receipt
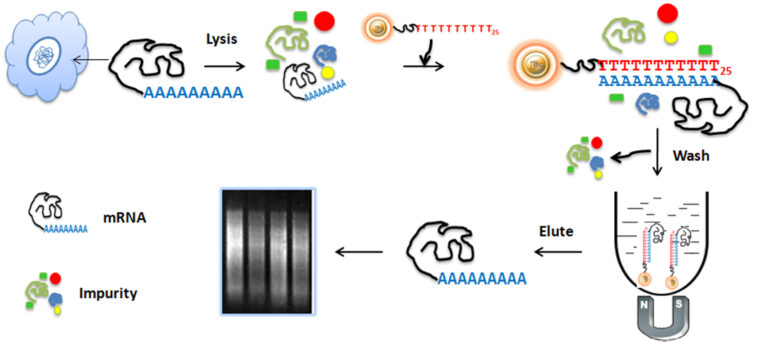
The BcMag™ Quick mRNA Purification Kit has been meticulously crafted to extract poly(A)+ RNA from cells and tissue without any use of phenol or other chemical solvents. The mechanism is quite intricate – Oligo d(T)30 is attached to 1µm paramagnetic beads, which then serve as a stable foundation for direct binding of poly(A)+ RNA. This technique offers the flexibility of manual processing for multiple samples, while still being able to adapt for automated high-throughput applications. What’s more, the magnetic separation technology enables the elution of undamaged mRNA in small amounts, eliminating the need for poly(A)+ transcripts to precipitate in the eluent. Astonishingly, it takes less than an hour to generate fully representative, intact poly(A)+ RNA of the original sample’s mRNA population.
The workflow involved in this is a thing of beauty. The beads are combined with cell lysates and incubated while continuously rotating for an adequate period. During mixing, the beads stay suspended in the sample solution, allowing the mRNA to bind to oligo-dT magnetic beads. Ribosomal RNA and other small RNA molecules, such as transfer RNA, microRNA, small nucleolar RNA, and small cytoplasmic RNA, don’t stick to the beads and are swiftly eliminated. After the incubation process, only the polyadenylated RNA species (mRNA) are collected and separated from the sample using a magnet rack. Finally, the ultrapure mRNAs are eluted and ready for use in downstream applications.
●
Suitable for high-throughput automated applications
●
No phenol or other organic solvents are required.
●
Precipitation of poly(A)+ transcripts in eluent is not required.
●
Genomic DNA contamination is minimal.
●
Beads that are reusable
●
Get pure mRNA in less than an hour.
●
Save time. Directly isolate mRNA from cell lysates and tissue homogenates (isolation of total RNA not required).
●
Prepare mRNA suitable for practically every downstream application.
●
Save time. Isolate mRNA directly from cell lysates and tissue homogenates (isolation of total RNA not required).
●
Allow for a diverse set of samples. Use both small and large-scale mRNA preparations.
●
cDNA synthesis
●
cDNA library creation
●
RT-PCR, quantitative RT-PCR
●
RPA (Ribonuclease Protection Assay)
●
Subtractive hybridization
●
Dot/slot hybridization
Notes
●
Different samples contain a variable amount of total RNA. At the same time, only 1-5% of total RNA is mRNA.
●
Each user must estimate the yield of RNA from the experimental sample. If the yield of RNA is much lower than expected, the following causes should be considered.
Materials Required
Buffer
●
BcMag oligo-d(T)25 magnetic beads: 10 mg/ml in 50 mM Tris-HC, pH 7.5, 0.5 M NaCl, 1mM EDTA, 0.1% NaN3
●
1x mRNA Binding Buffer: 0.1 M HEPES, pH 7.5, 0.5 M LiCl, 10 mM EDTA, 1% SDS, 10mM Dithiothreitol (DTT)
●
Washing Buffer I: 10mM HEPES, pH 7.5, 0.15 M LiCl, 1mM ETDA, 0.1% SDS
●
Washing Buffer II: 10mM HEPES, pH 7.5, 0.15 M LiCl, 1mM ETDA
●
Elution Buffer: 10mM HEPES, pH 7.5
Equipment
●
Magnetic Rack (for manual operation)
Based on sample volume, the user can choose one of the following magnetic Racks:
BcMag™ Rack-2 for holding two individual 1.5 ml centrifuge tubes (Bioclone, Cat. No. MS-01)
BcMag™ Rack-6 for holding six individual 1.5 ml centrifuge tubes (Bioclone, Cat. No. MS-02)
BcMag™ Rack-24 for holding twenty-four individual 1.5-2.0 ml centrifuge tubes (Bioclone, Cat. No. MS-03)
BcMag™Rack-50 for holding one 50 ml centrifuge tube, one 15 ml centrifuge tube, and four individual 1.5 ml centrifuge tubes (Bioclone, Cat. No. MS-04)
BcMag™ Rack-96 for holding a 96 ELISA plate or PCR plate (Cat. No. MS-05).
For larger scale purification, Ceramic magnets Block for large scale purification (6 in x 4 in x 1 in block ferrite magnet, Applied Magnets, Cat. No. CERAMIC-B8).
●
Corning 430825 cell culture flask for large-scale purification (Cole-Parmer, Cat. No. EW-01936-22).
●
Mini BlotBoy 3D Rocker, fixed speed, small 10″ x 7.5″ platform w/ flat mat (Benchmark Scientific, Inc. Cat. No. B3D1008) or compatible
Note: Contaminating RNAse or improper handling quickly degrades RNA molecules during purification. For the best results, follow the following guidelines.
●
Try to use fresh samples. Samples should be quickly frozen in liquid nitrogen and stored at –80ºC.
●
All glassware and plastic containers should be incubated in 1.0 M NaOH for four h to eliminate RNAse.
●
And then thoroughly rinsed with ultrapure water. However, disposable plastic tubes or other containers that have been pre-sterilized can be used directly.
●
Fresh ultrapure water (Milli-Q grade) can directly be used to prepare solutions.
●
Always wear disposable gloves and change them frequently.
A. Sample Treatment
Note: Add 1M Dithiothreitol (DTT) to 1x mRNA Binding Buffer to make a final concentration of 10 mM of DTT.
A1. Animal/Plant tissues
1.
Wholly and quickly homogenize the desired amount of plant or animal tissue in liquid nitrogen.
2.
Collect and transfer the frozen powder to a fresh tube. Add 1ml 1x mRNA Binding Buffer per 100 mg tissue, mix well by inverting the tube several times and incubate at room temperature for 5 min with rotational mixing.
3.
Reduce solution viscosity by shearing DNA through a syringe with an 18-gauge needle by sucking it in and out several times.
4.
Centrifuge at 14,000 rpm for 5 min at room temperature and transfer all the supernatant to a fresh tube.
5.
The supernatant is ready for mRNA purification or can be stored at –80ºC for future use.
A2. Cell suspension
1.
Pellet cells by centrifugation at 4,000 rpm for 5 min at room temperature and discard the supernatant. Add 1 ml 1x mRNA Binding Buffer per 5 x 106 – 10 x 106 animal or plant cells.
2.
Lyse cells by pipetting several times until the solution becomes viscous. Reduce solution viscosity by shearing DNA through a syringe with an 18-gauge needle by sucking it in and out several times.
3.
Centrifuge at 140 00 rpm for 5 min at room temperature and carefully transfer supernatant to a fresh tube. The supernatant is ready for mRNA purification or can be stored at –80ºC for future use.
A3. Yeast and Fungi
1.
Harvest yeast or fungi cells from the desired amount of culture by centrifugation at 14,000 rpm for 6 min at room temperature and remove all the supernatant. Quickly flash-freeze sample by immersion in liquid nitrogen bath.
2.
Add 1 ml 1x mRNA Binding Buffer per 1.5 ml culture and let the sample thaw on ice. Add 70 μl glass beads (sigma Cat. No. G-8772) per 1.5 ml culture and place them in a vortex mixer for 3 min.
3.
Centrifuge at 14,000 rpm for 5 min at room temperature and transfer all the supernatant to a fresh tube.
4.
Reduce solution viscosity by shearing DNA through a syringe with an 18-gauge needle by sucking it in and out several times.
5.
Centrifuge at 14,000 rpm for 5 min at room temperature and carefully transfer supernatant to a fresh tube. The supernatant is ready for mRNA purification or can be stored at –80ºC for future use.
A4. Total RNA
1.
The concentration of total RNA should be adjusted to ~1 μg/μl with ultrapure water. Divide the RNA sample into aliquots of 100 μl per tube, heat at 70°C for 5 min to destroy the RNA secondary structure, and immediately chill on ice.
2.
Add 1ml 1x Binding buffer to the RNA solution.
B. mRNA Purification
1.
Gently shake the bottle of BcMag•mRNA beads until the magnetic beads are completely suspended and transfer the desired amount of the beads to a fresh tube.
2.
Place the tube in a magnetic Rack and wait for 1-3 min until the supernatant becomes clear.
3.
Remove and discard the supernatant. Suspend the beads with 0.5 volumes of 1x Binding Buffer.
4.
Combine the magnetic beads with the pre-treated sample or the sample stored at 80ºC as described above.
Note: Samples previously stored at –80ºC should first be thawed on ice.
5.
Remove the tube from the magnetic Rack, mix the beads very well and incubate at room temperature for 10 min with rotational mixing.
6.
Place the tube in the magnetic Rack, wait 1 min and discard the supernatant. Resuspend the beads with four volumes of 1x Washing buffer I.
7.
Repeat (step 5) once.
8.
Place the tube in the magnetic Rack, wait 1 min and discard the supernatant. Resuspend the beads with four volumes of 1x Washing buffer II.
9.
Repeat (step 8) once.
10.
Place the tube in the magnetic Rack, wait 1min and discard the supernatant completely.
11.
Carefully transfer the mRNA-containing supernatant to a fresh tube. Heat the tube at 65ºC for 3 min and immediately separate the beads from the released mRNA by the magnetic Rack for 2 min. Add 20μl elution buffer, remove the tube from the magnetic Rack, and mix well.
Note:
●
If mRNA is used for cDNA synthesis, the magnetic beads should be washed with 100 μl 1x Reverse Transcription buffer one time after step 9. The mRNA can be eluted by adding the desired amount of 1x Reverse Transcription buffer, heated at 65ºC for 3 min and collected by magnetic Rack. or the bonded mRNA
●
Beads can be directly used in reverse transcription reactions.
●
Or the mRNA/bead complex can also be directly used for cDNA synthesis without elution after step 9.
●
The mRNA/bead complex should be washed one time with 100 μl 1x Reverse Transcription buffer, and then suspend mRNA/bead complex in the desired amount of 1x Reverse transcription buffer.
C. Storage
The eluted mRNA solution can be stored at –20˚C for a short time. For long-term storage, the mRNA solution should be stored in 75% ethanol at -80ºC.
D. Troubleshooting
Problem
The yield of purified mRNA is low or undetectable.
Probable Cause
Sample homogenization is incomplete.
Suggestion
Problem
The yield of purified mRNA is low or undetectable.
Probable Cause
mRNA is not completely dissolved.
Suggestion
Problem
The yield of purified mRNA is low or undetectable.
Probable Cause
Suggestion
Problem
The yield of purified mRNA is low or undetectable.
Probable Cause
mRNA degradation
Suggestion
Problem
Ribosomal RNA (rRNA) contamination
Probable Cause
The purified mRNA from some samples may carry over a small amount of rRNA. For some applications, such as Northern blots, the contamination will not significantly affect the experimental results. However, if the mRNA is used.
Suggestion
For cDNA library construction, rRNA should be eliminated from the mRNA solution. Usually, rRNA can be removed by repeating the same purification method used for the total RNA once more.
Problem
Probable Cause
Suggestion
The yield of purified mRNA is low or undetectable.
Sample homogenization is incomplete.
mRNA is not completely dissolved.
The pre-treated sample is too viscous.
If the pre-treated sample is too viscous, it will influence the efficiency of the beads’ binding to mRNA.
mRNA degradation
Ribosomal RNA (rRNA) contamination
The purified mRNA from some samples may carry over a small amount of rRNA. For some applications, such as Northern blots, the contamination will not significantly affect the experimental results. However, if the mRNA is used.
For cDNA library construction, rRNA should be eliminated from the mRNA solution. Usually, rRNA can be removed by repeating the same purification method used for the total RNA once more.
1.
Ferrier DC, Shaver MP, Hands PJW. Micro- and nano-structure-based oligonucleotide sensors. Biosens Bioelectron. 2015 Jun 15;68:798-810.
2.
Sethi D, Gandhi RP, Kuma P, Gupta KC. Chemical strategies for immobilization of oligonucleotides. Biotechnol J. 2009 Nov;4(11):1513-29.
3.
Zuo P, Ye BC. A novel immobilization strategy using oligonucleotide as linker for small molecule microarrays construction. Biosens Bioelectron. 2008 Jun 15;23(11):1694-700.
Magnetic Beads Make Things Simple