- +1 858 909 0079
- +1 858 909 0057
- [email protected]
- +1 858 909 0079
- [email protected]

Products
Components
Shipping conditions: At ambient temperature
Components
BcMag™ U-DNA Beads
10x Lysis Buffer (100mM Tris-HCl, PH 9.0)
Proteinase K
DTT
Proteinase K Suspension Buffer
Storage
4°C
4°C
-20°C
-20°C
4°C
Cat. No. AQ101 (50 Preps)
2.5 ml
0.6 ml
12.5 mg
15.4 mg
1.0 ml
Cat. No. AQ102 (100 Preps)
5.0 ml
1.2 ml
25 mg
30.8 mg
2.0 ml
Shipping conditions: At ambient temperature
Handling and Storage: Store the kit components according to the table Above on arrival.
Genomic variation analysis is critical to plant genetics and crop improvement efforts. Plant scientists require tools to address various concerns such as disease resistance, transgenic seed generation, yield improvement, population genetics, and plant physiology. High-quality DNA extraction from leaf, seed, root, or other plant material is required for reproducible DNA analysis using quantitative PCR, genotyping by sequencing (GBS), or next-generation sequencing (NGS).
Standard procedures for extracting plant DNA can be exceedingly Time consuming and labor intensive. Specific applications like plant genotyping demand a lengthy DNA extraction method. For example, traditional plant DNA extractions need a 4-hour to overnight digestion with proteinase K, followed by phenol: chloroform extraction and alcohol precipitation. Other techniques, such as silica bind and elute kits, have recently become available. Those techniques are based on positive chromatography purifying selection. High chaotropic salt concentrations, such as guanidine hydrochloride, bind DNA to silica. The silica column or beads is washed with a salt/ethanol solution after nucleic acid binding to eliminate additional biomolecules from the sample. Finally, the column or beads is eluted using Tris elution buffer or water to remove the pure DNA. Such bind-wash-elute procedures are time-consuming, requiring multiple washing and spinning steps. Repetitive spin steps can cause considerable DNA loss (20-40%) and shearing. Furthermore, chaotropic salts and other impurities can easily pass through the eluted DNA or RNA, compromising ultimate purity and quantification as well as downstream enzymatic activities like PCR.
BcMag™ One-Step Plant DNA Purification Kit allows rapid, column-free extraction of genomic DNA from various plant sample sources, including leaves, stems, buds, flowers, fruit, seeds, etc. The kit uses our unique proprietary magnetic beads and buffers to efficiently lyse cells and remove all impurities simultaneously in an aqueous buffer, leaving the DNA untouched. The procedure employs mild lysis conditions, avoiding harsh conditions such as alkaline lysis and toxic chemicals for lysing cells to maintain DNA integrity and the time-consuming cleanup of organic solvent from the sample. Furthermore, the magnetic beads eliminate PCR inhibitors (Fig.1) from samples in a single step without DNA extraction. It increases DNA integrity, boosts nucleic acid yields, and minimizes DNA loss caused by typical DNA purification techniques’ time-consuming “bind-wash-elute” procedure. Following sample lysis, the straightforward one-step purification technique enables simultaneous processing of >96 samples and produces pure DNA in less than 30 minutes. Purified genomic DNA has the highest integrity and can be used in various downstream applications such as qPCR.

1.
Use bead beating to disrupt the sample in a bead beater.
2.
Centrifuge and transfer the supernatant to a new tube.
3.
Mix the samples with the magnetic beads and proteinase K and heat to lyse the cells.
4.
Vortex the beads to capture PCR inhibitors.
5.
Remove the beads with a magnet.
6.
Aspirate the supernatant containing the pure ready-to-use DNA.
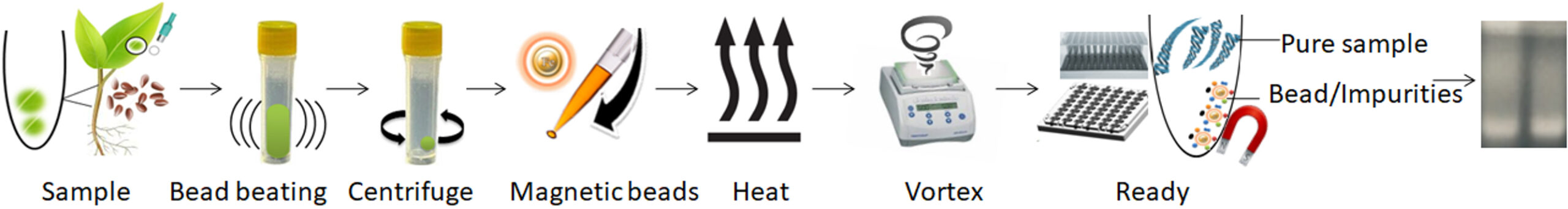
The purified DNA is ready for downstream applications, such as PCR, qPCR, single-nucleotide polymorphism (SNP), genotyping, genotyping by sequencing (GBS), next-generation sequencing (NGS), etc.
●
Rapid and efficient purification protocol: without prior DNA isolation for subsequent use in direct workflows, No liquid transfer, and One-tube.
●
Ultrafast: Process 96 samples in less than an hour.
●
Highest nucleic acids recovery rates: Minimal loss of DNA during extraction.
●
●
Cost-effective: Eliminates columns, filters, laborious repeat pipetting, and organic reagents.
●
High throughput: Compatible with many different automated liquid handling systems.
Magnetic Beads Make Things Simple