- +1 858 909 0079
- +1 858 909 0057
- [email protected]
- +1 858 909 0079
- [email protected]

Next-Generation Sequencing (NGS) is a cutting-edge technology that has revolutionized the field of genomics. Also known as massively parallel sequencing, it is a high-throughput, quick, and scalable method that can replace first-generation Sanger sequencing. NGS enables the simultaneous interrogation of hundreds to thousands of genes in multiple samples and the discovery and analysis of various types of genomic features in a single sequencing run, ranging from single nucleotide variants (SNVs) to copy number and structural variants.
To ensure high-quality sequencing data, it is essential to have a proper NGS workflow in place. The NGS workflow consists of four critical steps: nucleic acid extraction, library preparation, sequencing, and analysis. Each step is crucial to the success of your experiment, and the decisions you make at each step can have a significant impact on the quality of your data.
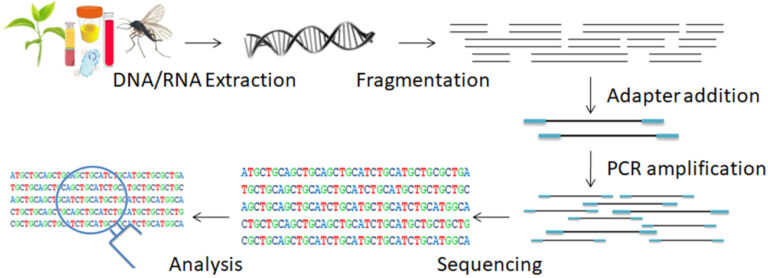
The extraction of nucleic acids for sequencing is the first step in the NGS process. DNA, total RNA, mRNA, or chromatin can be extracted from a wide variety of biological samples, including tumor biopsy, early embryos, embryonic tissues, and circulating DNA. However, most existing procedures are time-consuming, costly, cumbersome, and require vast amounts of DNA, making them unsuitable for samples with limited amounts of starting material. Moreover, contaminants frequently act as inhibitors of enzymatic reactions by obstructing or degrading the template sample, competing with ions in solution, or even denaturing the enzymes. Some contaminants are inherent to the sample type, such as polysaccharide complexes or tannic acids in plants, hemoglobin in the blood, or urea in urine samples.
The extraction technique can also introduce contaminants. Reagents often used in “manual” extraction techniques include chaotropic salts, alcohols, salts, and phenol: chloroform. The extraction of a sufficient amount of DNA from the sample input without removing disrupting inhibitors can be challenging. Carry-over contamination during sequencing runs can cause considerable errors.
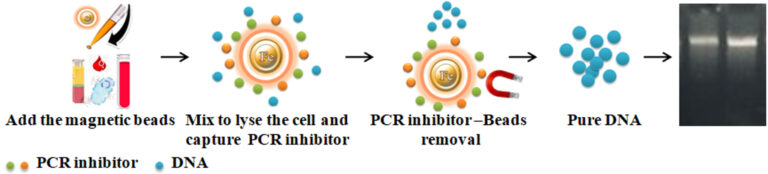
To address this issue, Bioclone has developed a novel DNA purification system, the BcMag™ One-step DNA Purification System, which uses negative chromatography magnetic beads to deliver high-quality and superior DNA yield from most trace DNA samples. The magnetic beads with proprietary surface chemistry function simultaneously to lyse cells and capture the PCR inhibitors once mixed with the sample. The magnetic beads-PCR inhibitor complex is then magnetically removed by a magnet, while the pure DNA remaining in the solution is ready for downstream STR analysis.

1.
Add functional magnetic beads to the sample.
2.
Mix the samples with the magnetic beads and heat to lyse the cells.
3.
Vortex for 5 minutes for the beads to capture the PCR inhibitors.
4.
Remove the beads with a magnet.
5.
Aspirate the supernatant containing the pure ready-to-use DNA.
●
Rapid and efficient purification protocol without prior DNA isolation for subsequent use in direct amp workflows, No liquid transfer, and One-tube.
●
Ultrafast: Process 96 samples in less than an hour.
●
Higher purity and DNA yield with minimal contamination with RNA from various trace samples.
●
●
Cost-effective: Eliminates columns, filters, laborious repeat pipetting, and organic reagents.
●
High throughput: Compatible with many different automated liquid handling systems.
Learn More
Several processing procedures are required to prepare a library. Physical shearing or enzyme digestion is used to fragment DNA (or cDNA) samples into a small, homogeneous piece of DNA. The resultant DNA fragments are first end-polished and subsequently ligated to sequencing adapters. These adapter sequences are used to amplify the insert DNA by PCR to generate a fragment library.

Next Generation Sequencing (NGS) libraries require high-quality nucleic acid inputs in variable volumes, concentrations, and sizes depending on the library preparation procedures and sequencing platforms utilized. Regardless of these differences, traditional hands-on approaches such as magnetic beads, columns, or agarose gel electrophoresis are typically used as part of the entire technique for labs attempting to assess the quality of determinants in this manner. These techniques are classified into two types based on their function:
Size Selection: Using size selection beads or BcMag™ One-step NGS Cleanup kit to removes undesired nucleic acid fragments or library molecules that are larger or smaller than a specific size range that is ideal for the downstream sequencing platform.
Sample Cleanup: Use BcMag™ One-step NGS Cleanup kit to removes sequencing adaptors or PCR primers, dNTPs, enzymes, or undesirable buffer formulations from the sample.
Current technologies and chemistries have been in use for several years for the goals indicated above; nevertheless, they are used at the expense of performance and convenience. Many library preparation processes require repeated purifications, which might result in DNA loss. With each purification step, current methods can result in up to a 30%-50% loss. That may eventually need more starting material, which may not be possible with limited, valuable samples, or the addition of more PCR cycles, which may result in sequencing bias. Reproducibility between samples is a daily difficulty that is frequently mentioned as a potential area for development in size selection.
It is now possible to efficiently and precisely purify dsDNA for NGS, PCR, and general molecular biology applications. BcMag™ One-Step NGS Cleanup Kit is specially designed for ultrafast and efficient purification of post PCR reaction or potential replacement of size selection procedure after adaptor addition. The protocol is not only straightforward (one tube and one step) but also very flexible in removing different size DNA fragments by adjusting processing time, buffer’s pH, and detergent concentration (table1). The magnetic Beads are added directly to the finished PCR reactions or other DNA reactions and mixed by a vortex mixer or pipetting to capture and remove the impurities (e.g., excess primer, dimer, adapter, salt, detergent, dNTPs, and enzyme). After mixing, the beads are magnetically removed, while the supernatant contains the purified and ready-to-run products. The beads enable 96 samples to be processed simultaneously in less than 10 minutes. In just 1 minute, the purified DNA is ready for downstream applications, such as Sanger Sequencing, Restriction Digestion, Cloning, SNP Detection, or Library Preparation for NGS.
●
Simple protocol: No liquid transfer, One-tube, One-step
●
Ultrafast: One-minute protocol
●
Higher purity and recovery > 90% DNA
●
●
Cost-effective: Eliminates columns, filters, laborious repeat pipetting, and ethanol
●
High-throughput: Compatible with many different automated liquid handling systems
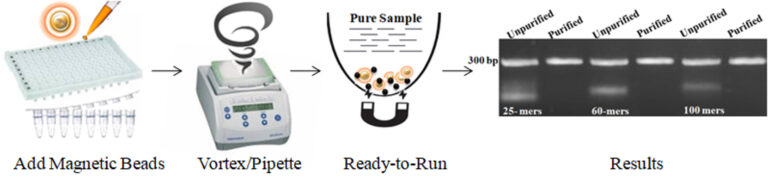
Table 1 – DNA Fragment Removal
Table 1 – DNA Fragment Removal
DNA
Buffer
+ 0.1%
Triton x-100
pH7.5
– 0.1%
Triton x-100
pH7.5
+ 0.1%
Triton x-100
pH 8.0
– 0.1%
Triton x-100
pH 8.0
+ 0.1%
Triton x-100
pH 8.8
– 0.1%
Triton x-100
pH 8.8
dsDNA
(100 bp)
No Removal
Removal
Removal
Removal
No Removal
Removal
dsDNA
(150 bp)
No Removal
Removal
No Removal
Removal
No Removal
Removal
dsDNA
(200 bp)
No Removal
Removal
No Removal
Removal
No Removal
Removal
dsDNA
(300 bp)
No Removal
No Removal
No Removal
No Removal
No Removal
No Removal
ssDNA
100 mer
Removal
Removal
Removal
Removal
Removal
Removal
Please Note:
dsDNA – Double-Stranded DNA; ssDNA – Single-stranded DNA
The assay was done by using the following conditions:
1. 10 mM Tris-HCl with or without 0.1% triton (final concentration) and three different: pH 7.5, pH 8.0 and pH 8.8
Learn More
Following clonal amplification, an NGS platform is used for parallel sequencing. The majority of sequencing technologies are based on the detection of nucleotides incorporated by the DNA polymerase and are thus classified as “sequencing by synthesis” (SBS) approaches. Several NGS technologies have been developed, including pyrosequencing, sequencing by ligation (SOLiD), sequencing by synthesis (SBS – Illumina), and Ion Torrent sequencing.
Each NGS experiment generates a massive amount of complex data made up of short DNA reads. Although each technology platform has its algorithms and data analysis tools, they share a typical analysis ‘pipeline’ and employ common metrics to assess the quality of NGS data sets.
1.
Dorado G, Gálvez S, Rosales TE, Vásquez VF, Hernández P. Analyzing Modern Biomolecules: The Revolution of Nucleic-Acid Sequencing – Review. Biomolecules. 2021 Jul 28;11(8):1111.
2.
Beck TF, Mullikin JC; NISC Comparative Sequencing Program, Biesecker LG. Systematic Evaluation of Sanger Validation of Next-Generation Sequencing Variants. Clin Chem. 2016 Apr;62(4):647-54. doi: 10.1373/clinchem.2015.249623. Epub 2016 Feb 4.
3.
Rocca MS, Ferrarini M, Msaki A, Vinanzi C, Ghezzi M, De Rocco Ponce M, Foresta C, Ferlin A. Comparison of NGS panel and Sanger sequencing for genotyping CAG repeats in the AR gene. Mol Genet Genomic Med. 2020 Jun;8(6):e1207.
4.
Ke R, Mignardi M, Hauling T, Nilsson M. Fourth Generation of Next-Generation Sequencing Technologies: Promise and Consequences. Hum Mutat. 2016 Dec;37(12):1363-1367.
5.
Werner T. Next generation sequencing in functional genomics. Brief Bioinform. 2010 Sep;11(5):499-511.
Magnetic Beads Make Things Simple