- +1 858 909 0079
- +1 858 909 0057
- [email protected]
- +1 858 909 0079
- [email protected]

Components
Shipping conditions: At ambient temperature
Components
BcMag™ U-DNA Beads
10x Lysis Buffer (100mM Tris-HCl, PH 9.0)
Proteinase K
DTT
Proteinase K Suspension Buffer
Storage
4°C
4°C
-20°C
-20°C
4°C
Cat. No. AE101 (50 Preps)
2.5 ml
5.0 ml
12.5 mg
15.4 mg
1.25 ml
Cat. No. AE102 (100 Preps)
5.0 ml
10 ml
25 mg
30.8 mg
2.5 ml
Shipping conditions: At ambient temperature
Handling and Storage: Store the kit components according to the table Above on arrival.
The extraction of genomic DNA from bacteria has long been a challenging task, requiring a laborious and time-consuming process that often involves the use of columns and toxic chemicals. But fear not, a novel and groundbreaking approach has been developed to streamline this process and revolutionize the field of DNA purification.
The single-step purification protocol for bacterial DNA is an incredibly innovative and efficient method that eliminates the need for columns, resulting in rapid extraction of high-quality genomic DNA from an incredibly diverse range of microorganisms. Whether you’re dealing with Gram-positive or Gram-negative bacteria, this system has got you covered.
It uses a proprietary combination of magnetic beads and buffers that work together to enable gentle cell lysis and simultaneous removal of contaminants. This approach ensures that the DNA remains intact, eliminating the need for harsher methods that often involve toxic chemicals or alkaline lysis.
With the inclusion of magnetic beads, this system also facilitates the removal of PCR inhibitors from samples in one easy step, which can dramatically increase DNA yield, maintain DNA integrity, and reduce the loss of DNA due to the laborious and time-consuming “bind-wash-elute” method used in conventional DNA purification methods.
Following sample lysis, this simple one-step purification technique can process up to 96 samples simultaneously and produce pure DNA within a brief period of less than 30 minutes. The genomic DNA purified by this method has exceptional quality and can be used in a wide variety of downstream applications, including qPCR, with high accuracy and sensitivity.
1.
Use bead beating to disrupt the sample in a bead beater.
2.
Centrifuge and transfer the supernatant to a new tube.
3.
Mix the samples with the magnetic beads and proteinase K and heat to lyse the cells.
4.
Vortex the beads to capture PCR inhibitors.

5.
Remove the beads with a magnet.
6.
Aspirate supernatant containing the pure ready-to-use DNA.
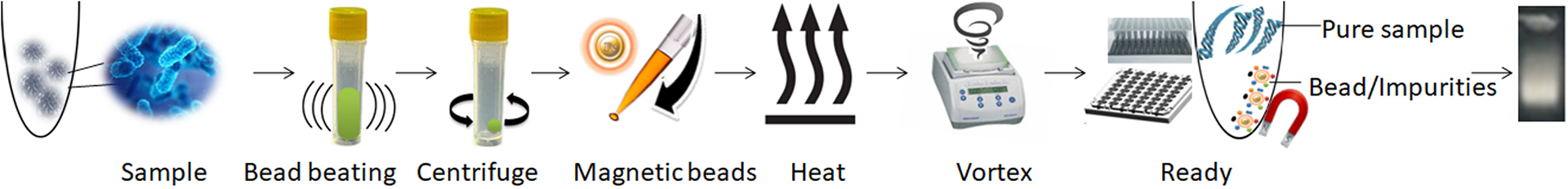
The One-Step Bacteria DNA Purification Kit offers several advantages over traditional DNA purification methods. It provides rapid DNA extraction, allowing for the simultaneous processing of more than 96 samples in less than 30 minutes. This is beneficial for high-throughput applications, where a quick and efficient DNA extraction method is necessary. The kit uses unique proprietary magnetic beads and buffers to lyse cells and remove impurities simultaneously in an aqueous buffer, ensuring efficient DNA purification without the need for additional purification steps.
One of the significant advantages of the One-Step Bacteria DNA Purification Kit is that it maintains high DNA integrity and yield. It does not use harsh conditions that may lead to DNA degradation, ensuring that high-quality DNA yields are suitable for downstream applications such as qPCR. The kit also eliminates PCR inhibitors from samples in a single step without DNA extraction, further enhancing DNA integrity and yield.
Another benefit of the One-Step Bacteria DNA Purification Kit is that it eliminates the need for columns, reducing the cost and time of the purification process. This makes it a more affordable and efficient option for DNA extraction, without compromising the quality and yield of the extracted DNA.
The purified DNA is ready for downstream applications, such as PCR, qPCR, single-nucleotide polymorphism (SNP), short tandem repeat (STR) genotyping, genotyping, next-generation sequencing (NGS, veterinary genotyping, etc.
The following protocol is an example. The protocol can be scaled up or down as needed.
Notes
●
DNA Yield: Varies (depends on sample size and type)
●
DNA Size: Varies (depends on the quality of starting material
●
Since there is no concentration step in the protocol, the concentration of the nucleic acid depends on the quality and quantity of the sample used.
●
Quantification of the nucleic acids: Use only fluorescence methods such as qPCR, Qubit, and Pico Green.
●
●
For long-term storage, store the extracted nucleic acids at -20°C.
Materials Required by the User
Item
Magnetic Rack for centrifuge tube
** Based on sample volume, the user can choose one of the following magnetic Racks
Source
• BcMag™ Rack-2 for holding two individual 1.5 ml centrifuge tubes (Bioclone, Cat. No. MS-01)
• BcMag™ Rack-6 for holding six individual 1.5 ml centrifuge tubes (Bioclone, Cat. No. MS-02)
• BcMag™ Rack-24 for holding twenty-four individual 1.5-2.0 ml centrifuge tubes (Bioclone, Cat. No. MS-03)
• BcMag™ Rack-50 for holding one 50 ml centrifuge tube, one 15 ml centrifuge tube, and four individual 1.5 ml centrifuge tubes (Bioclone, Cat. No. MS-04)
Item
BcMag™ 96-well Plate Magnetic Rack.
Source
• BcMa™ 96-well Plate Magnetic Rack (side-pull) compatible with 96-well PCR plate and 96-well microplate or other compatible Racks (Bioclone, Cat. No. MS-06)
Item
Adjustable Single and Multichannel pipettes
Item
Centrifuge with swinging bucket
Addition items are required if using 96-well PCR plates / tubes
Vortex Mixer
** The user can also use other compatible vortex mixers. However, the Time and speed should be optimized, and the mixer should be: Orbit ≥1.5 mm-4 mm, Speed ≥ 2000 rpm
Eppendorf™ MixMate™
Eppendorf, Cat. No. 5353000529
Tube Holder PCR 96
Eppendorf, Cat. No. 022674005
Tube Holder 1.5/2.0 mL, for 24 × 1.5 mL or 2.0 mL
Eppendorf, Cat. No. 022674048
Smart Mixer, Multi Shaker
BenchTop Lab Systems, Cat. No. 5353000529
1.5/2.0 mL centrifuge tube
96-well PCR Plates or 8-Strip PCR Tubes
PCR plates/tubes
** IMPORTANT! If using other tubes or PCR plates, make sure that the well diameter at the bottom of the conical section of PCR Tubes or PCR plates must be ≥2.5mm.
Items
Magnetic Rack for centrifuge tube
** Based on sample volume, the user can choose one of the following magnetic Racks
Source
●
BcMag™ Rack-2 for holding two individual 1.5 ml centrifuge tubes (Bioclone, Cat. No. MS-01)
●
BcMag™ Rack-6 for holding six individual 1.5 ml centrifuge tubes (Bioclone, Cat. No. MS-02)
●
BcMag™ Rack-24 for holding twenty-four individual 1.5-2.0 ml centrifuge tubes (Bioclone, Cat. No. MS-03)
●
BcMag™ Rack-50 for holding one 50 ml centrifuge tube, one 15 ml centrifuge tube, and four individual 1.5 ml centrifuge tubes (Bioclone, Cat. No. MS-04)
BcMag™ 96-well Plate Magnetic Rack
●
BcMa™ 96-well Plate Magnetic Rack (side-pull) compatible with 96-well PCR plate and 96-well microplate or other compatible Racks (Bioclone, Cat. No. MS-06)
Adjustable Single and Multichannel pipettes
Centrifuge with swinging bucket
Addition items are required if using 96-well PCR plates/tubes
Vortex Mixer
** The user can also use other compatible vortex mixers. However, the Time and speed should be optimized, and the mixer should be: Orbit ≥1.5 mm-4 mm, Speed ≥ 2000 rpm
Eppendorf™ MixMate™
Tube Holder PCR 96
Tube Holder 1.5/2.0 mL, for 24 × 1.5 mL or 2.0 mL
Smart Mixer, Multi Shaker
Eppendorf, Cat. No. 5353000529
Eppendorf, Cat. No. 022674005
Eppendorf, Cat. No. 022674048
BenchTop Lab Systems, Cat. No. 5353000529
Eppendorf™ MixMate™
Tube Holder PCR 96
Tube Holder 1.5/2.0 mL, for 24 × 1.5 mL or 2.0 mL
Eppendorf, Cat. No. 5353000529
Eppendorf, Cat. No. 022674005
BenchTop Lab Systems, Cat. No. 5353000529
1.5/2.0 mL Centrifuge Tube
96-well PCR Plates or 8-Strip PCR Tubes
PCR plates/tubes
! IMPORTANT ! If using other tubes or PCR plates, ensure that the well diameter at the bottom of the conical section of PCR Tubes or PCR plates must be ≥2.5mm.
A. Sample Preparation
Sample material lysis and disruption
We recommend the Bead Beating method for sample material lysis and disruption. Bead beating is an effective method that disrupts nearly any biological sample to release the DNA, RNA, and proteins found within the cells. Disrupting the sample material must be complete to obtain the best DNA yields. Samples are placed in tubes with grinding beads and subjected to high-energy mixing. The beads make contact with the sample, eventually breaking it down at the cellular level and releasing subcellular contents. Typically, the samples are centrifuged, and the lysate retrieved from above the beads.
Instrument
●
Beadblaster 24 microtube homogenizer (Benchmark Scientific, Catalog No. D2400)
●
Vortex-Genie 2 (Fisher Scientific, Catalog No.50-728-03)
●
MN Bead Tube Holder for Vortex-Genie 2 (Takara, Catalog No. 740469)
●
FastPrep® homogenizer (MP Biomedicals)
General recommendations for properly homogenizing samples
●
Do not overfill tubes. The combined volume of the sample, beads, and buffer should never be more than half the size of the tube. Less is more.
●
The volume of the sample and buffer should be double that of the beads.
●
Detergents should be used sparingly since the foaming will obstruct the movement of the beads.
●
If sample warming is an issue, process the sample in short bursts.
●
Solid tissues should never be more than 1/20th the volume of the disruption tube, i.e., no more than 100 mg tissue in a 2 ml tube.
The user must select the optimal disruption period based on sample type, sample amount, vortex frequency, and liquid volume in the tube according to the manufacturer’s instructions.
●
When possible and appropriate, cut the sample into small pieces to facilitate processing.
●
Avoid overloading the sample tube to allow efficient mixing of Lysis Mix with the sample.
Sample
Example sample input
Bacteria samples (Fresh or frozen)
Method 1
1.
Add 1 mg-2 mg microbial pellet (wet weight) to a 2 ml lysis tube containing appropriate grinding beads. Add 750 µl** of 1x lysis buffer to the tube and cap tightly. (Note: Add 3 µl of 10 mM DTT to 100 µl of 1x lysis buffer immediately before use.) Follow the manufacturer’s instructions to process the sample.
2.
Centrifuge at ≥12,000 x g for 5 minutes.
3.
Transfer 10 µL supernatant to each well of PCR plate or PCR tube (using 96-well PCR plates/tubes method).
B. Premix Beads Solution Preparation
! IMPORTANT !
1.
Before pipetting, shake or Vortex the bottle to completely resuspend the Magnetic Beads.
2.
Do not allow the magnetic beads to sit for more than 2 minutes before dispensing.
3.
Proteinase K preparation: Provide protease K as lyophilized powder and dissolve at a 20 mg/ml concentration in Proteinase K Suspension Buffer. For example, 12.5 mg dissolved in 625 µl of Proteinase K Suspension Buffer. Divide the stock solution into small aliquots and store at -20°C. Each aliquot can be thawed and refrozen several times but should then be discarded.
4.
DTT solution preparation: Provide DTT as powder and dissolve at a concentration of 1M in dH2O. For example, 15.4 mg dissolved in 100µl dH2O. It is stable for years at -20°C. Prepare in small aliquots, thaw it on ice, and use and discard. Store them in the dark (wrapped in aluminum foil) at -20°C. Do not autoclave DTT or solutions containing it. Avoid multiple freeze-thaw cycles.
5.
Dilute DTT to a concentration of 10 mM from stock with dH2O and use it immediately. Discard unused DTT solution.
6.
Prepare a fresh Master Mix following Table 2 for the number of samples to be processed, plus 10% more (e.g., if you have 10 samples, prepare Master Mix for 11). Add the following components to the reservoir.
Table 2. Premix Beads solution
Components
BcMag™ U-DNA Beads
10x Lysis Buffer
Proteinase K (20mg/ml)
DTT (10 mM)
Sample
DH2O
Total
One Well ( 100 μL Reaction Volume)
50 μl
10 μl
12.5 μl
3 μl
x
x
100 μl
Components
C. Isolation Procedure
! IMPORTANT !
●
Pipet up and down premix beads solution in a reagent reservoir until the solution is homogeneous before dispensing.
●
Do not allow the magnetic beads to sit for more than 5 minutes before dispensing.
1.
Transfer 100µl premix beads solution to the sample to a new well of 96well PCR plate or 0.2ml PCR tube and add the sample.
2.
Mix the sample well by Vortex or pipetting.
3.
Place the PCR plate/tube into a thermocycler and incubate at:
a.
65°C for 15 minutes
b.
80°C for 10 minutes
4.
Remove the PCR plate/tube from the thermocycler and then mix the sample with beads by slowly pipetting up and down 20-25 times, or Vortex the sample at 2000 rpm for 5 minutes (see picture).

5.
Centrifuge at 3500 rpm for 5 minutes.
6.
Place the sample plate/ tube on the magnetic separation plate for 30 seconds or until the solution is clear.
7.
Transfer the supernatant to a clean plate /tube while the sample plate remains on the magnetic separation plate. The sample is ready for downstream applications. Using 1-5 ul in a 25μl of qPCR reaction.
D. Troubleshooting
Problem
Low DNA/RNA Recovery
Probable Cause
Poor starting sample material
Suggestion
Problem
Ct Value Delays
Probable Cause
Too many PCR inhibitors in the sample.
Suggestion
Problem
Ct Value Delays
Probable Cause
Recovery DNA is so low.
Suggestion
Problem
Probable Cause
Suggestion
Low DNA/RNA Recovery
Poor starting sample material
Ct Value Delays
Too many PCR inhibitors in the sample.
Recovery DNA is so low.
Magnetic Beads Make Things Simple