- +1 858 909 0079
- +1 858 909 0057
- [email protected]
- +1 858 909 0079
- [email protected]

Polymerase chain reaction (PCR) is becoming the standard method for detecting and characterizing microorganisms and genetic markers in a wide range of sample types. However, the approach is susceptible to inhibitory chemicals present in the sample and may decrease the assay’s sensitivity or potentially result in false-negative results.
PCR inhibitors are any factors that prevent nucleic acid amplification via the polymerase chain reaction (PCR). When enough copies of DNA are present, PCR inhibition is the most prevalent reason for amplification failure. Many distinct inhibitory compounds can be found in a single matrix, while the same inhibitors can exit in multiple matrices. Organic substances such as bile salts, urea, phenol, ethanol, polysaccharides, sodium dodecyl sulphate (SDS), humic acids, tannic acid, melanin, and various proteins such as collagen, myoglobin, hemoglobin, lactoferrin, immunoglobin G (IgG), and proteinases
are among these inhibitors. Organic and inorganic compounds, whether liquid or solid, can act as PCR inhibitors. Calcium ions are an example of an inorganic material that inhibits PCR.
PCR inhibitors exit in a wide range of biological materials (organs, blood, body fluids, and so on), environmental samples (water, soil, air, and so on), and food (meat, milk, fruits, vegetables, seafood, etc.). Furthermore, inhibiting compounds may be mistakenly introduced during transportation, sample processing (e.g., pre-concentration techniques), or nucleic acid extraction.
PCR inhibitors can disrupt various steps of PCR analysis. Specific PCR inhibitors may interfere with the annealing of the primers to the DNA template. Template RNA or DNA can be degraded or modified by nucleases. The presence of polysaccharides may limit the ability to resuspend precipitated RNA. Reverse transcription can be blocked, for example, by the enzyme’s direct contact with melanin.
Many PCR inhibitors directly or indirectly target the DNA polymerase. Polymerase activity may be inhibited by calcium, collagen, haematin, and tannic acid. Protease or detergent involved in the process can degrade the polymerase.
High calcium concentrations may cause the DNA polymerase to compete with magnesium for binding, and complexing agents, such as tannic acid, deplete magnesium. In both circumstances, magnesium is no longer available as a cofactor for the polymerase, resulting in a diminished activity. Interference with fluorescent probes or increased background fluorescence reflects other PCR inhibitors’ action modes that reduce sensitivity in real-time PCR assays.
It uses control to determine the quantity of inhibition that occurs in a reaction by introducing a known amount of a template to the investigated reaction mixture (based on the sample under analysis). The level of inhibition can be analyzed by comparing the amplification of this template in the mixture to the amplification seen in a separate experiment using the identical template in the absence of inhibitors. This technique will underestimate the inhibition as it applies to the investigated sequence (s). Of course, suppose any inhibition in the sample-derived reaction mixture is sequence-specific.
PCR inhibitors exist in a variety of biological materials (organs, blood, body fluids, etc.), environmental samples (water, soil, air, etc.), and food (meat, milk, fruits, vegetables, seafood, etc.).Furthermore, inhibitors may be mistakenly introduced during transport., sample processing (e.g., pre-concentration procedures), or nucleic acid extraction. Table1 shows some examples of matrices and their specific inhibitors.
Sources
Blood, Muscle tissues
Inhibitors
Heparin, Hemoglobin, Immunoglobulins, Proteases, Nucleases, Acyclovir, Hormones, IgG, Lactoferrin, Myoglobin, Collagen
Milk
Proteases, Calcium ions
Stool
Plants
Soil
Sample preparation
DNA purification
Dirty plasticware
Bile salts, Complex polysaccharides, Lipids, Urate
Pectin, Polyphenols, Polysaccharides, Xylan, Chlorophyll
Fulmic acids, Humic acids, Humic material, Metal ions, Polyphenol
KCl, SDS, Xylene
Chaotropic salts, Ethanol, Isopropanol, Phenol, Sodium acetate
Protease, Nucleases
PCR Inhibitor
In blood, serum, or plasma samples, molecules such as IgG, hemoglobin, and lactoferrin are PCR inhibitors. Many of these inhibitors influence RNA directly rather than the reaction’s enzymes. Anticoagulants, such as heparin may also hinder PCR. Furthermore, hormones and antiviral medications such as acyclovir can impact amplification. The most critical component in urine samples is urea, which can cause polymerase breakdown. Bile salts, urea, glycolipids, hemoglobin, and heparin are examples of inhibitors. Stool and fecal samples contain polysaccharides or chlorophyll derived from herbs and vegetables.
There are numerous PCR inhibitors found in the diet. Food ingredients such as fats, glycogen, polysaccharides, minerals, and enzymes may hinder the PCR. In milk samples, PCR inhibition is mostly determined by calcium concentration and plasmin. Polysaccharides appear to be primarily responsible for PCR suppression in seafood. Plants include numerous compounds, such as polysaccharides, polyphenols, pectin, and xylan, which can be co-extracted and impede PCR. Environmental samples can be highly diverse and come from various compartments such as soil, water, or air. This comprehensive range results in numerous PCR inhibitors, including those already mentioned.
Pathogens are routinely detected using a wide range of water samples. Fats, proteins, polyphenols, and heavy metals can exist in sewage sludge, and wastewater contains polysaccharides, metal ions, and RNases, all of which are common PCR inhibitors of environmental materials. Furthermore, humic and fulminic acids, which block PCR even at low doses, may be present in dead biomass and soil. Large volumes of water are typically concentrated in very small proportions to identify trace concentrations of pathogens. However, this frequently leads to contemporaneous concentrations of the various inhibitors and more significant interference with PCR. Similar issues may arise while collecting air samples, often accomplished by passing vast volumes of air through multiple filters or other binding material. As a result, airborne inhibitory components are frequently concentrated, resulting in PCR failure.
During sample processing or nucleic acid extraction, inhibitors may be introduced to the sample. It comprises glove powder, various detergents, organic compounds, sarkosyl, ethanol, isopropyl alcohol, or phenol. These compounds may be required for effective cell lysis or the preparation of pure nucleic acids, but at specific quantities, they may also block PCR. Non-ionic detergents (e.g., Nonidet P-40, Tween 20, Triton X-100, and N-octyl glucoside) block PCR only at relatively high doses, whereas ionic are significantly inhibitory.
There are several approaches for the removal of inhibitors or the reduction of their effects.
1.
Dilute the sample extracts containing PCR inhibitors. It is simple, but the diluted DNA may not be enough for successful DNA amplification. A decrease in sensitivity accompanies dilution.
2.
Chelex such as Chelex1-100 and Phenol–Chloroform-based protocols. Both methods are inefficient for removing most of the PCR inhibitors since Chelex can only remove some divalent ions. In contrast, the Phenol–Chloroform method can only deplete lipids and proteins.
3.
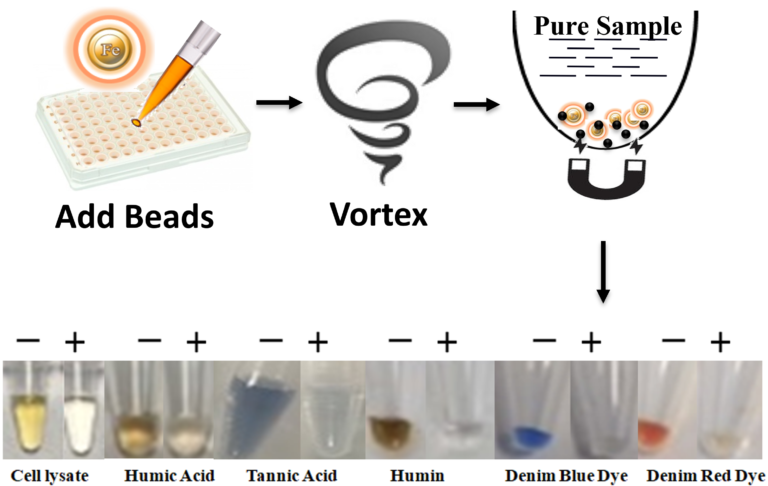
Column chromatography such as Sephacryl S-400, Sephadex G-200, and silica-based spin-column or magnetic beads effectively remove some of the inhibitors, but the process is labor-intensive and time-consuming.
4.
Choose an appropriate enzyme
The DNA polymerase must show that specific Taq DNA polymerase mutations can overcome inhibition by blood, plasma, hemoglobin, lactoferrin, serum IgG, soil extracts, and humic acids.
In summary, although these methods can remove some PCR inhibitors, however, practically they have the following limitations such as:
1.
Less efficient, time-consuming, or labor-intensive.
2.
Potential loss of DNA sample during processing.
3.
Chaotropic salt and ethanol are potentially carried over into the eluted DNA.
4.
These are not suitable for high-throughput processing and automation.
Explore Product
1.
Abbaszadegan, M., Huber, M.S., Gerba, C.P. and Pepper, I.L. (1993) Detection of enteroviruses in groundwater with the polymerase chain reaction. Appl Environ Microbiol 59, 1318–1324.
2.
Abbaszadegan, M., Stewart, P. and LeChevallier, M. (1999) A strategy for detection of viruses in groundwater by PCR. Appl Environ Microbiol 65, 444–449.
3.
Abolmaaty, A., Gu, W., Witkowsky, R. and Levin, R.E. (2007) The use of activated charcoal for the removal of PCR inhibitors from oyster samples. J Microbiol Methods 68, 349–352.
4.
Alaeddini, R. (2011) Forensic implications of PCR inhibition – a review. Forensic Sci Int Genet 6, 297–305.
5.
Baar, C., d’Abbadie, M., Vaisman, A., Arana, M.E., Hofreiter, M., Woodgate, R., Kunkel, T.A. and Holliger, P. (2011) Molecular breeding of polymerases for resistance to environmental inhibitors. Nucleic Acids Res 39, e51.
Get the Latest News and Updates by Email
6393 Nancy Ridge Dr. Suite A
San Diego, CA 92121 USA
Fax: +1-858-909-0057
Get the Latest News and Updates by Email
© 2023 Bioclone Inc. All Rights Reserved.